Abstract
Background: Mutations in the myeloid transcription factor RUNX1 are frequently detected in AML and MDS. Differences have been described regarding additional cytogenetic and molecular genetic abnormalities, however the molecular factors responsible for progression of RUNX1 mutated MDS to AML are yet not fully elucidated.
Aim: Analysis of genetic (1) similarities and (2) differences of RUNX1 mutated AML and MDS cases, (3) identification of factors potentially causing disease progression from RUNX1 mutated MDS to AML.
Methods: Whole-genome sequencing (WGS) was performed for 773 AML and 747 MDS patients (median coverage 100x). 151bp paired-end reads were generated on NovaSeq 6000 and HiSeqX machines (Illumina, San Diego, CA). As no sample specific normal tissue was available, a so-called Tumor/Unmatched normal (TUN) workflow was used to reduce technical artefacts and germline calls. All reported p-values are two-sided and were considered significant at p<0.05.
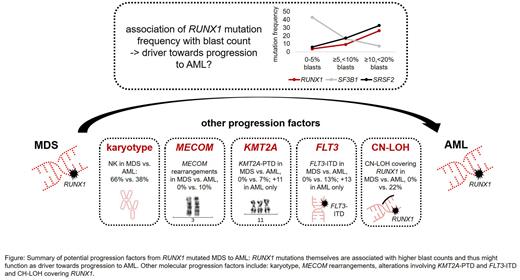
Results: Mutations in RUNX1 (RUNX1mut) were detected at comparable frequencies in AML and MDS (AML: 97/773 cases, 13%; MDS: 71/747, 10%). In MDS, an association of the RUNX1mut frequency with higher blast counts was observed (0-5% blasts: 15/419 with RUNX1mut, 4%; ≥5,<10%: 16/175, 9%; ≥10,<20%: 40/153, 26%). However, blast count did not influence overall survival (OS) in RUNX1mut subgroups (0-5% blasts: 18 months (m); ≥5,<10%: 25 m; ≥10,<20%: 18 m; AML: 21 m). The variant allele frequency (VAF) of RUNX1mut differed significantly dependent on the blast count (median VAF in cases <5% blasts: 0.22; ≥5%: 0.42; p<0.001). Regarding cytogenetic abnormalities, RUNX1mut MDS showed a higher frequency of normal karyotype (NK) compared to AML (66% vs. 38%, p<0.001) but similar frequencies of trisomies (18% vs. 22%) and complex karyotype (3% vs. 5%), although differences regarding the trisomy type were detected (MDS: only trisomy 8; AML: trisomies 8, 11 and 13). In 10% of AML cases with RUNX1mut a rearrangement involving MECOM was detected (5% GATA::MECOM, 5% other MECOM rearrangements), which was absent in MDS with RUNX1mut. Similarly, KMT2A-PTD and FLT3-ITD alterations were detected in AML but not in MDS with RUNX1mut (KMT2A-PTD: 7% in AML vs. 0% in MDS, p=0.021; FLT3-ITD: 13% vs. 0%, p=0.001). By contrast, TP53mut were found at comparably low frequencies (MDS: 4/71, 6%; AML: 3/97, 3%). For both AML and MDS, a strong association of RUNX1mut with mutations in spliceosome genes (SF3B1, SRSF2, U2AF1, ZRSR2) was detected (AML: 58/97 = 60% in RUNX1mut vs. 139/676 = 21% in RUNX1wild type (wt); p<0.001; MDS: 52/71 = 73% vs. 346/676 = 51%, p<0.001). However, while SRSF2mut in MDS were associated with increasing blast counts similar to RUNX1mut (0-5% blasts: 25/419 with SRSF2mut, 6%; ≥5,<10%: 130/175, 17%; ≥10,<20%: 50/153, 32%), SF3B1mut were negatively correlated to blast counts and thus to RUNX1mut (0-5% blasts: 179/419, 43%; ≥5,<10%: 28/175, 16%; ≥10,<20%: 11/153, 7%). In the total AML cohort, in 24/773 of cases a CN-LOH involving RUNX1 was detected, 21 of these also showed a RUNX1mut (22% (21/97) of RUNX1mut cases). By contrast, in the total MDS cohort in only 2/747 cases a CN-LOH covering RUNX1 was found, none of them harboring a RUNX1mut. The VAF of cases with RUNX1mut + CN-LOH (RUNX1mutLOH) was >0.5 for all cases and was significantly higher than the VAF of RUNX1mut cases without CN-LOH (0.89 (range 0.54-0.97) vs. 0.42 (range 0.05-0.85), p<0.001). Moreover, cases with RUNX1mutLOH were found to be significantly associated with FLT3-ITD (RUNX1mutLOH, 6/21 = 29%; RUNX1mut without CN-LOH, 7/76 = 9%; p=0.032).
Conclusions: (1) RUNX1mut occur at similar frequencies in AML and MDS and genetically show an MDS signature, reflected e.g. by association with mutations in spliceosome genes. OS is similar in all subgroups independent of blast counts. (2) In MDS, RUNX1mut are associated with higher blast counts, thus potentially functioning as driver towards progression to AML. (3) Additional cytogenetic/molecular genetic alterations that might function as progression factors from RUNX1mut MDS to AML include: (i) karyotype, especially MECOM rearrangements, trisomy 11 and trisomy 13 (ii) KMT2A-PTD, (iv) FLT3-ITD, (v) and CN-LOH covering RUNX1 (Figure). (4) By contrast, TP53mut do not seem to play a role in progression of RUNX1 mutated MDS to AML.
Disclosures
Stengel:MLL Munich Leukemia Laboratory: Current Employment. Meggendorfer:MLL Munich Leukemia Laboratory: Current Employment. Walter:MLL Munich Leukemia Laboratory: Current Employment. Baer:MLL Munich Leukemia Laboratory: Current Employment. Nadarajah:MLL Munich Leukemia Laboratory: Current Employment. Hutter:MLL Munich Leukemia Laboratory: Current Employment. Kern:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership. Haferlach:Munich Leukemia Laboratory: Current Employment, Other: Part ownership. Haferlach:MLL Munich Leukemia Laboratory: Current Employment, Other: Ownership.
Author notes
Asterisk with author names denotes non-ASH members.